Dr. Arjun Sharma
Dr. Mario Brameshuber
Biophysics Group, Institute Of Applied Physics, TU Wien, Austria
Background
The Biophysics Group at TU Wien is interested in super-resolution imaging in order to study the dynamics of biological systems such as T cells. One approach to this is cryo super-resolution imaging, which aims to produce highly-resolved images by imaging at extremely low cryogenic temperatures of -180 °C. At this temperature fluorophores are far more stable: while samples imaged at room temperature may bleach within minutes, the same sample would last hours at cryogenic temperatures, allowing for long exposures and high light intensities with little risk of bleaching the sample.
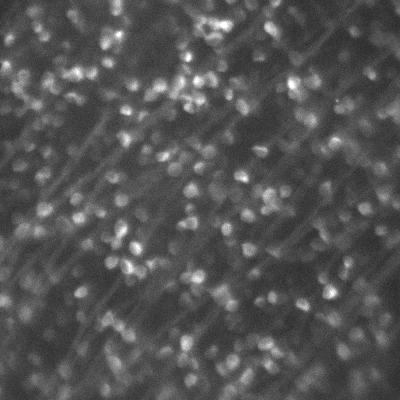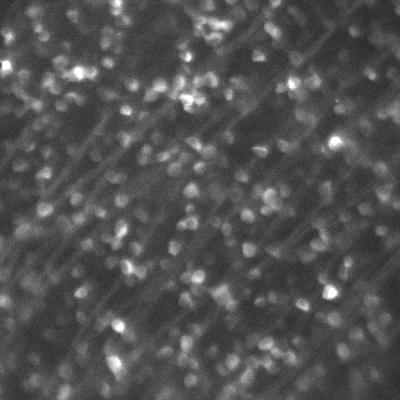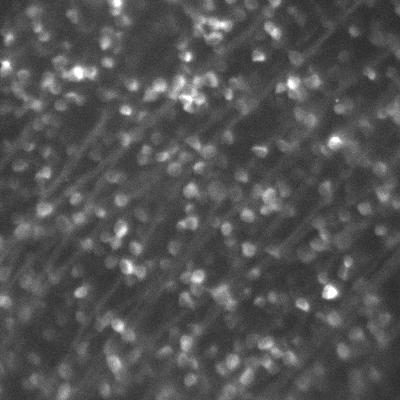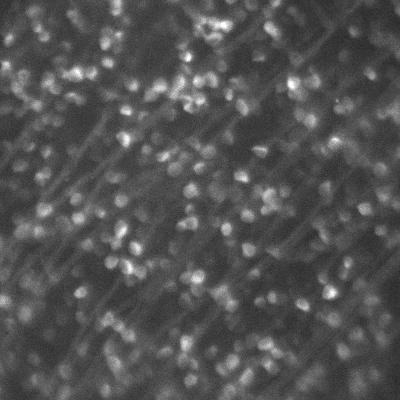
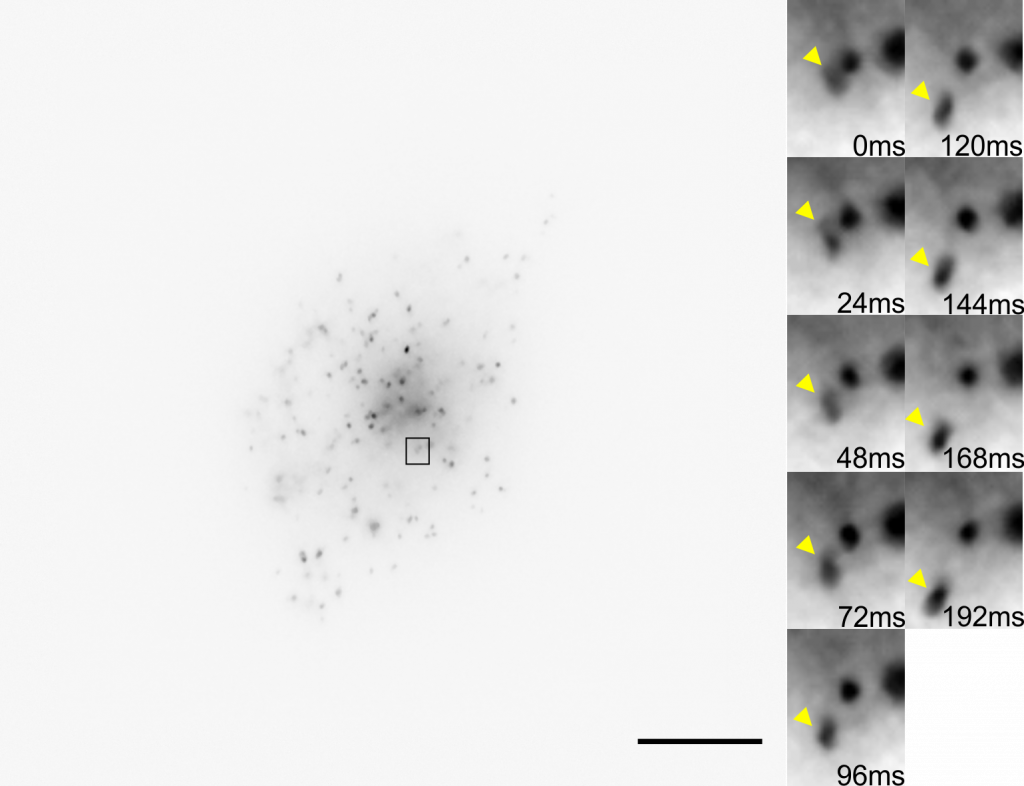
Dr. Arjun Sharma uses the cryogenic imaging system, and stated: “with blinking fluorophores and cryogenic temperatures, we can differentiate one molecule from another and increase the resolution up to 1 nm, this is our target.” This sub-cellular resolution allows for a detailed analysis of protein oligomers and aggregates within T cells and other samples.
Challenge
Due to the extremely low temperatures, the cryogenic system avoids standard imaging challenges concerned with photodamage and bleaching. However, the system introduces some unique challenges, as outlined by Dr. Sharma: “We are using a 60x low 0.7 NA air objective, the photon collection efficiency is not great so we really need a camera that is very sensitive and with minimum noise.” Only air objectives can be used with the cryogenic imaging system as water or oil would freeze, consequently, higher-NA immersion objectives cannot be used.
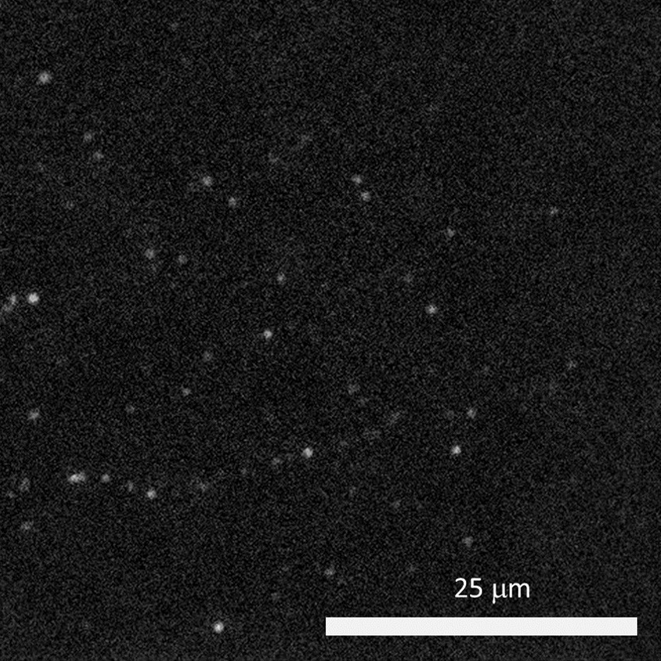
The cold also limits fluorophore blinking, meaning that a large field of view is needed in order to capture blinking and achieve super-resolution acquisition. The high exposure times mean that speed is not a factor, but a suitable camera should provide high sensitivity over a large field.
“The [Prime BSI] has high sensitivity and better signal to noise values than typical sCMOS cameras, and its large field of view allows us to capture up to four images in a single frame.”
Solution
The Prime BSI is an ideal fit for the cryogenic super-resolution system, with the balanced 6.5 μm pixel providing Nyquist-optimized resolutions at 60x. In addition, the low noise and 95% quantum efficiency result in a high signal to noise ratio when using the Prime BSI.
Dr. Sharma was also impressed by the Prime BSI: “From the specifications on your website it said your noise levels are better compared to normal CMOS camera and EMCCD, we checked that and also compared the noise level to another EMCCD we were using, and the images taken [with the Prime BSI] were much better.”