Prof. Geoffrey Murphy
Molecular & Behavioral Neuroscience Institute
University of Michigan
Background
Dr. Geoffrey Murphy, professor of physiology at the University of Michigan’s Molecular & Behavioral Neuroscience Institute studies the how the mammalian brain encodes, stores and retrieves information. Dr. Murphy explains, “We do a lot of mouse molecular genetics, in vitro neurophysiology, and mouse behavior. We are interested in the dendritic architecture, and the calcium signaling within the dendritic structure versus the somatic structure.”
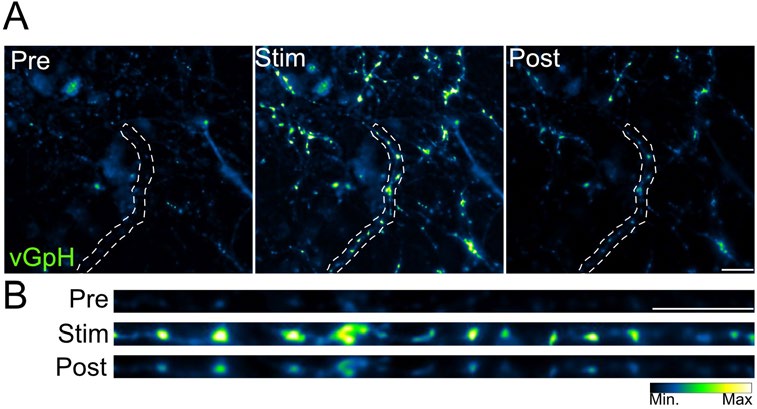
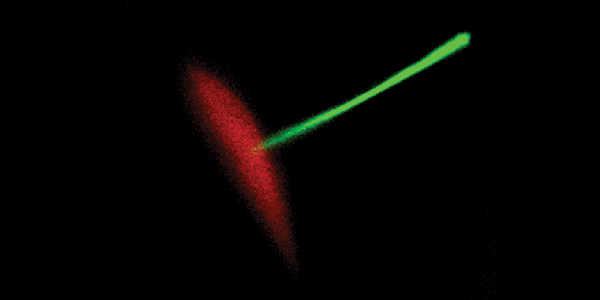
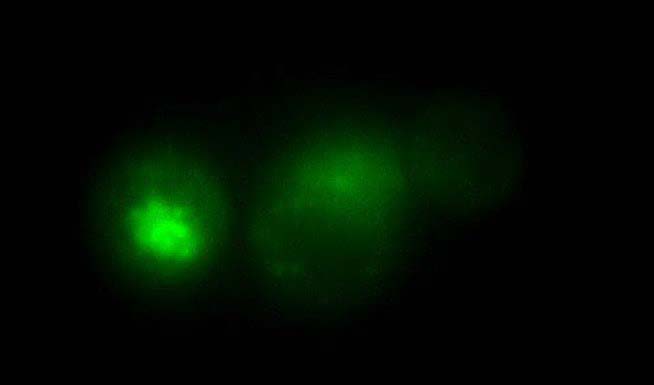
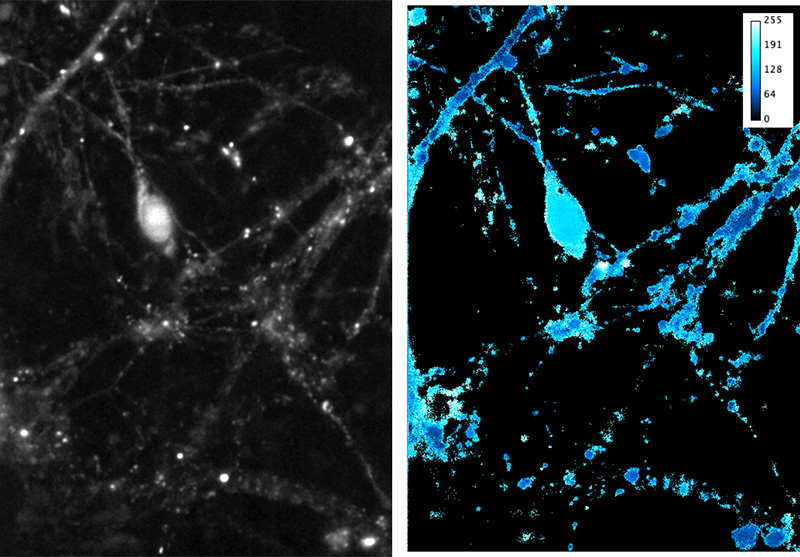
A: Images of fluorescent intensity before (Pre), during (Stim) and after electrical stimulation (100 pulses, 10 Hz) of neurons.
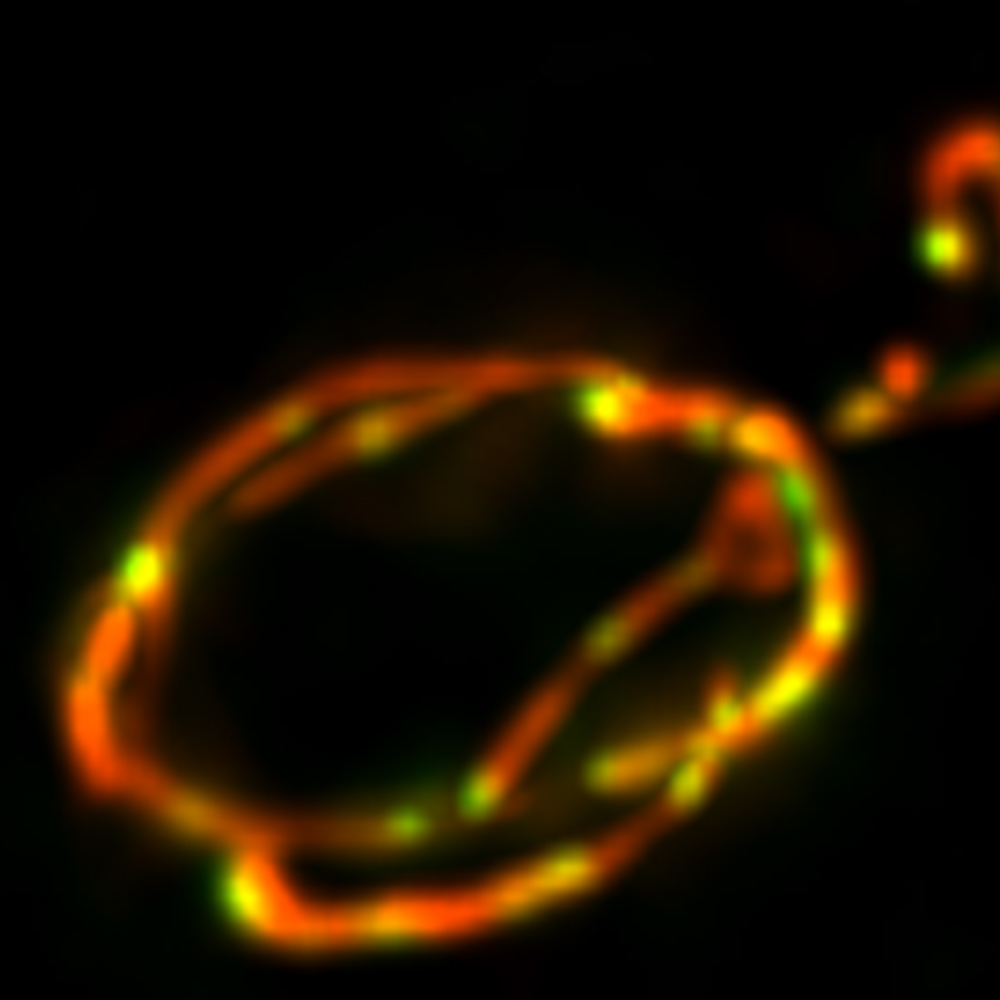
B: Enlarged view of an axonal segment (from outlined region in A).
Challenge
For many neuroscience researchers, speed, sensitivity, and resolution are all critical to visualizing small changes in calcium signals in different regions of the neuron. Dr. Murphy told us, “Being able to image at 100 Hz is very important to us. That allows us to get high resolution, rapid calcium signals.” Additionally, the samples to be imaged require a sensitive camera to visualize as the samples are often thick and can be troublesome to image through.
The Prime 95B [Scientific CMOS camera] allowed us to not only increase the frame rate we were using to acquire images, but we also achieved higher resolution. For us, that meant being able to look at subcellular structure in real time.
Solution
For Dr. Murphy, the 11µm pixel size and speed of the Prime 95B Scientific CMOS (sCMOS) provided a solution to the challenges of calcium imaging in neurons. Dr. Murphy told us, “Previously, we had been using cameras that had a lower frame rate and worse resolution, so using the Prime 95B allowed us to not only enhance the frame rate that we were using to acquire images, but get higher resolution too. For us, that meant being able to look at subcellular structures in real time so the speed is certainly something that we like. Also, it’s a really easy camera to use and it interfaces well with our preferred software application [for further analysis].”