Dr. Tom Lummen
D-BSSE Single Cell Facility, Department of Biosystems, ETH Zürich, Switzerland
Background
Dr. Tom Lummen is a microscopy engineer at ETH Zürich University, and spoke to us about the imaging facility he works with, “I’m part of the microscopy team that operates the imaging core facility, we provide 20 high-end automated microscopes for the imaging needs of the users of our department… we are always scouting for new or expanded functionalities for these systems.”
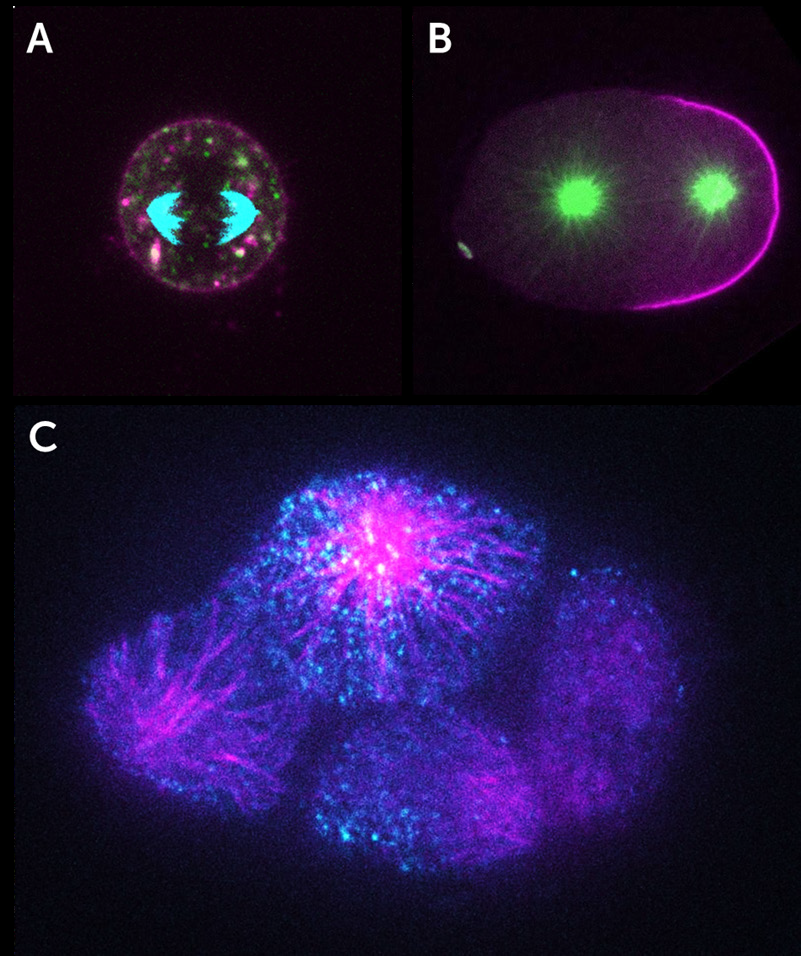
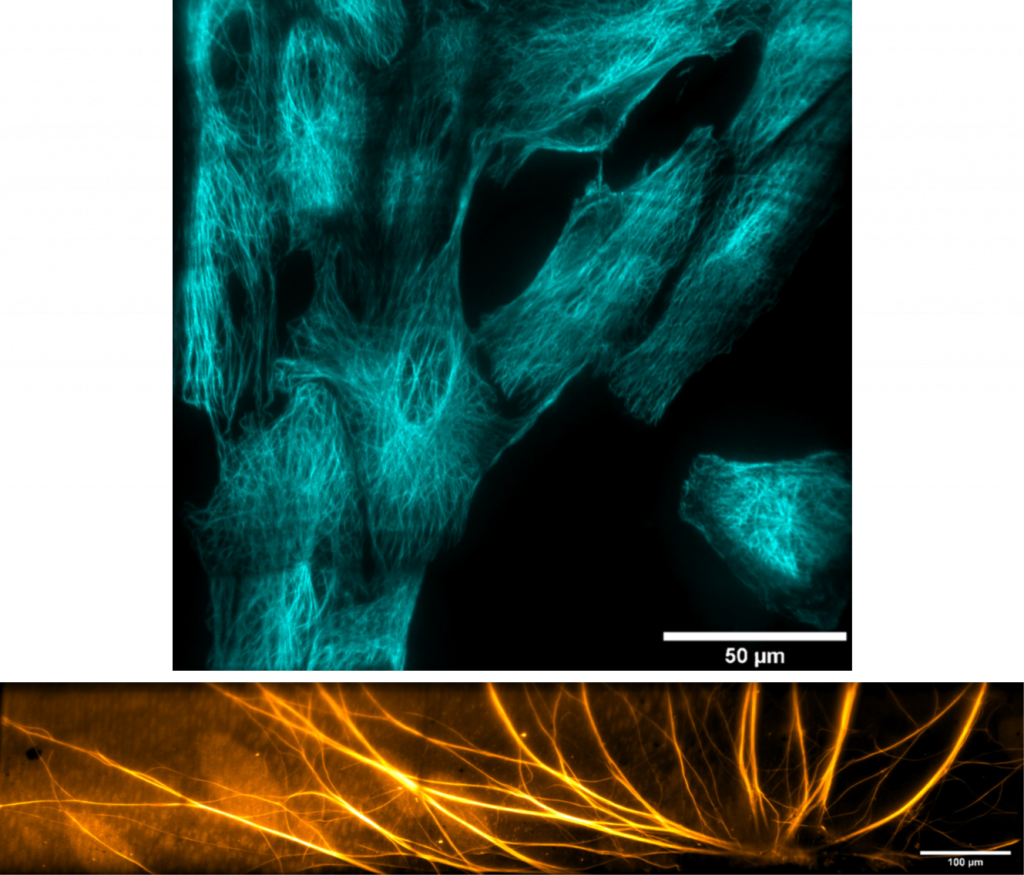
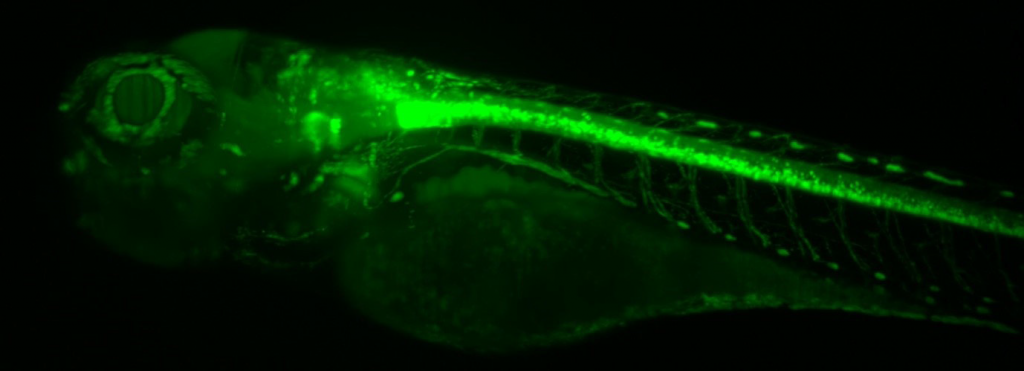
“We have a very broad array of imaging applications, these range from small subcellular features in live cells that have to be visualized on short timescales, all the way to the development of very big 3D organoids or tissue cultures that need to be monitored for weeks. The name of the game is flexibility and modularity!”
“Spinning disk confocal microscopy is becoming more in demand, as data acquisition needs are moving towards getting single-cell parameters but with very good statistics and/or time resolution. Our inverted spinning disk systems are geared towards live-cell observations for dynamic samples, but we have also seen an increase in demand for high 3D spatial throughput.”
Challenge
Spinning disk microscopy allows for fast imaging of live samples, but this technique can involve several challenges. Dr. Lummen described some of these challenges he has encountered, “Within microscopy there is always a trade-off when setting up a system, you can’t have all the specs at the same time. What we try to go for are dedicated systems, but we try to keep them as tuneable as possible.”
“We identified the need for a dedicated inverted spinning disk system that is geared towards high throughput observation, whether spatial or temporal. This is when we started to explore what camera technologies were out there.”
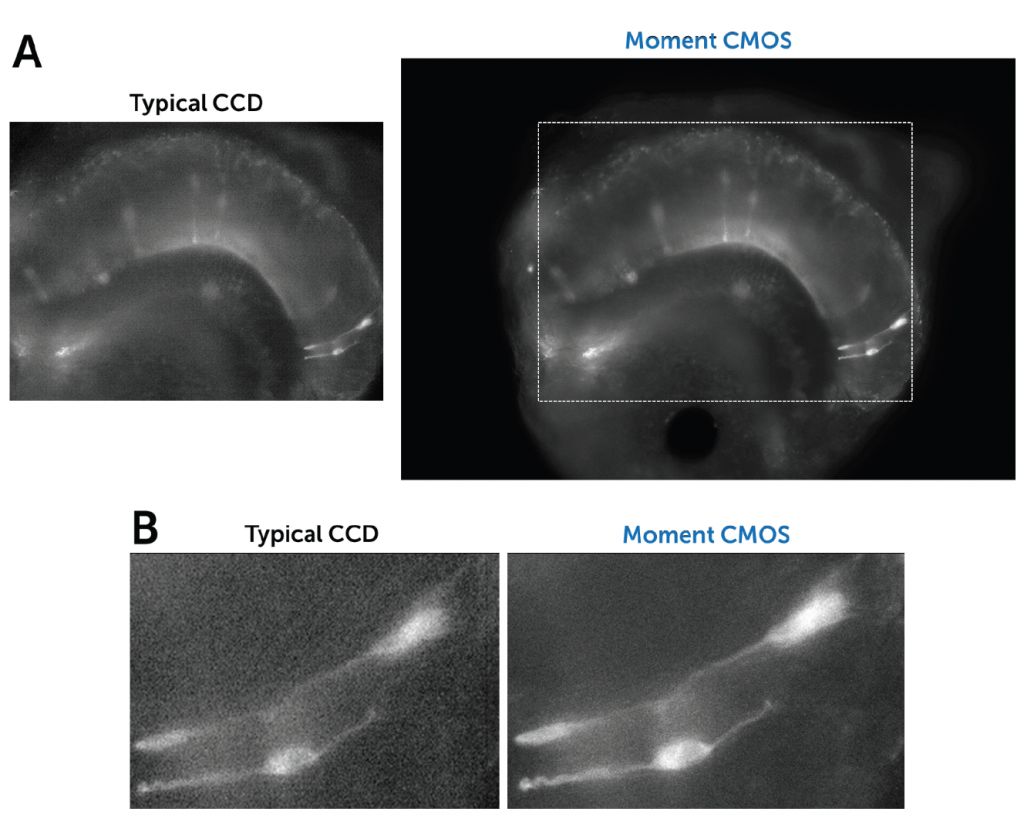
A suitable camera for high-throughput live-cell imaging at high speeds (to pair up with the speed of a spinning disk) would need to be highly flexible, have a large field of view (FOV), small pixels for high sampling resolution, high readout speeds and high sensitivity to detect the dynamics of moving samples.
The Kinetix is such an exciting product for us, because the large field of view allows us to really increase the useful data throughput, without compromising on readout speed or pixel size.
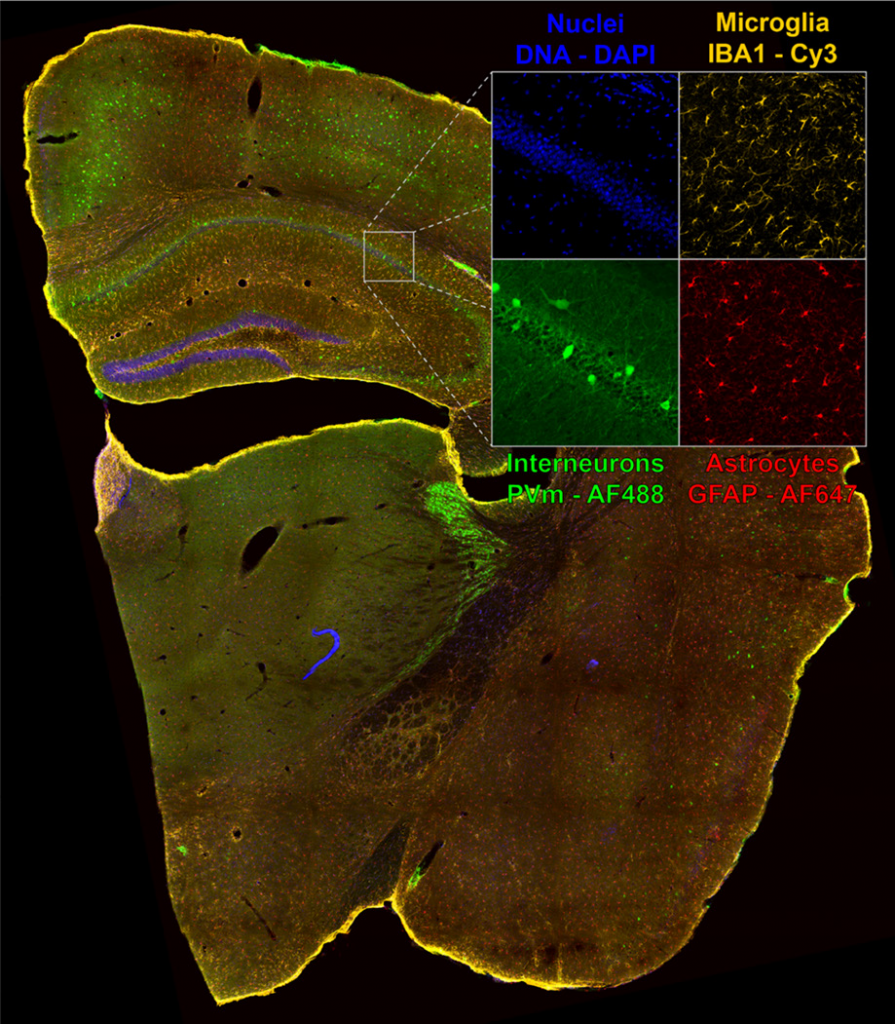
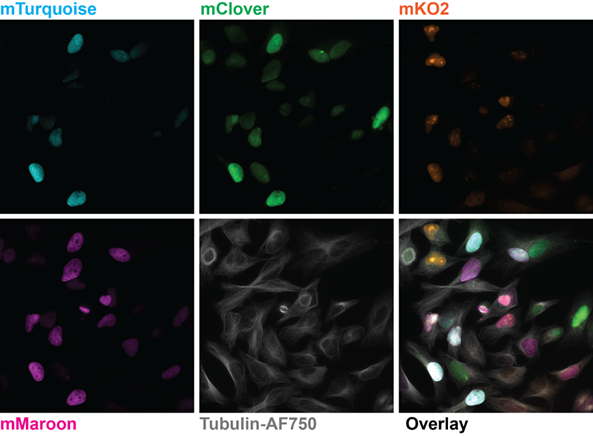
Solution
The Kinetix sCMOS was identified as a well-fitting solution for such dynamic live-cell spinning disk applications. Dr. Lummen shared his experiences with the Kinetix, “Imaging live-cell dynamics is one of the main reasons why the Kinetix is the right product for us, because the large field of view lets us image more cells in the same time span, which can proportionally increase the data throughput that we
can offer to our users.”
“We generally optimize our imaging setups on a per-experiment basis and tune for field-of-view, speed, resolution, sensitivity, or whichever trade-off thereof is most appropriate. The flexibility the Kinetix offers means that we’re not camera-restricted to one or two types of experiments; in principle, it can be configured for any of these optimized regimes.”
“The combination of a very large FOV, the small pixel size, and the high-speed specs of the Kinetix was really tempting to explore and we chose it for our new flexible high throughput system. It gives us a big boost to the FOV so we can get more cells per timepoint, or fewer tiles per composite large image. The combination with the large FOV of both the Nikon Ti2 and the CrestOptics X-Light v3 spinning disk confocal scanner let us configure a dual-camera large FOV setup. This enables us to further duplicate the data throughput or to perform true concurrent two-channel live-cell imaging in dynamic samples. In addition, with the Lumencor Celesta, this setup can do NIR imaging, adding an extra spectral channel we
can use.”
“For versatile systems, we often work with Nikon as the software is quite modular and open, and we can script more complex and conditional experiments…With the Kinetix directly coupled to the Nikon Ti2 we don’t see any hard vignetting at the corners of the chip and we can utilize the full 29 mm field of view, which is a big increase compared to our previous generation 19 mm sCMOS. The high sensitivity allows to run the disk at maximum speed, good for fluorescence observation of live cells.”

Reference
Bajar B., Lam A., Badiee R., Oh Y-H,, Chu J., Zhou X., Kim N., Kim B., Chung M., Yablonovitch A.,
Cruz B., Kulalert K., Tao J., Meyer T., Su X-D and Lin M (2016) Fluorescent indicators for
simultaneous reporting of all four cell cycle phases, Nat Methods 13, 993–996 https://
doi.org/10.1038/nmeth.4045