Introduction
There is currently a revolution in computational imaging, pairing the power of a cutting-edge scientific camera, such as the Prime Family of CMOS cameras from Teledyne Photometrics, with powerful graphics processors from the latest generation of GPUs. Machine learning, data processing, and the camera-PC interface are all going to help imaging evolve, through methods such as denoising and localization.
In this article, we explore our localization software for the Prime Family of sCMOS cameras, PrimeLocate™. Prime’s computational power matches the latest generation of GP GPU’s while making the technology easy to deploy and accessible by embedding computational power inside the camera.
Data Localization
An excess of image data may seem like an abundance of riches, except when the data does not contain useful information. Then it becomes data that must be stored, processed, and ultimately discarded, taking up processing capabilities and valuable digital storage space.
Advanced microscopy applications such as super-resolution and single-molecule often require localization, such as single-molecule localization microscopy (SMLM) for STORM or PALM imaging. With the high image quality of the Prime family, identifying and localizing these molecules is a simple matter, but the full image frame is transferred to the host for analysis when the molecules of interest may only occupy a tiny percentage of the frame. This results in a large amount of data transfer and processing time without adding useful information.
These issues can be solved using data localization, by recording data only from the relevant points of the camera sensor and ignoring the large areas of background without valuable data. This significantly reduces file sizes, storage strains, and allows for much faster processing.
The Prime family of sCMOS cameras from Teledyne Photometrics solves this localization problem at its source with PrimeLocate™, one of its advanced realtime processing features.
PrimeLocate™
When enabled, PrimeLocate automatically finds and transfers regions of interest to the host computer, either over PCIe or USB. In this way, massive amounts of data containing no useful information are eliminated from consideration. With no impact on camera acquisition speed, PrimeLocate™ can identify up to 512 of the brightest 3 x 3 pixel patches within each image and transmit only that portion of the image, reducing the data and processing requirements from ~6.7 Gbps down to ~100 Mbps. By transferring only the relevant raw data, users have the freedom to use their preferred localization algorithm to generate super-resolution images.
The PrimeLocate algorithm is most appropriate for single-molecule localization microscopy (SMLM) methods such as STORM, PALM and DNA-PAINT. The hallmark feature of SMLM is that sparse images of individual point emitters blink at random times during an image sequence. By finding the centroid of each emitter’s diffraction-limited spot in a given frame and combining the localization results from each frame, a super-resolution image of the original fluorescence can be reconstructed. This simplifies image capture, as well as reduces processing time and storage requirements, making your experiments more efficient!

The PrimeLocate algorithm scans each full image (or single ROI, if enabled) for the brightest local maxima within a 3 x 3 window. Each identified bright local maxima forms the center of a transmitted patch. The transmitted patch size is under user control and can vary from a radius of “1” to transmit a patch of 3 x 3 pixels, to a radius of “15” for transmitting a patch of 31 x 31 pixels. The number of transmitted patches can vary from one to a maximum of 512 patches per frame.
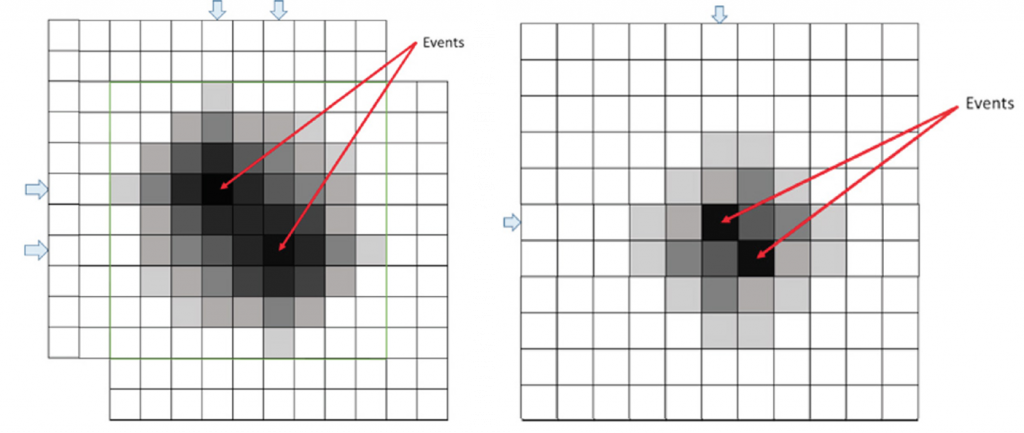
Using a small 3 x 3 pixel search window ensures that when two fluorescence events happen in close proximity, two or more patches are transmitted to capture both events, as shown in Figure 2. The 3 x 3 exclusion region discounts two or more events occurring within a radius of two pixels as being independent, as events this close together can be captured within a single, transmitted patch.
PrimeLocate always transmits the predetermined number of patches per frame, regardless of the number of fluorescence events present. For example, when using the maximum 512 regions, after patches containing true fluorescence events are transmitted, the remaining patches will simply contain the brightest noise peaks from the image. In this way, a consistent amount of data is transmitted per frame. These patches will be low-intensity compared to the brightest events and can be ignored. Should more than 512 events occur within a single frame, those events will be lost.
PrimeLocate does not attempt to localize events to a precision better than a pixel, as this step is expected to be performed on the host. Leaving this step to the host allows the choice of localization algorithm and configuration. The data transmitted to the host includes the patch data itself as well as the patch’s (X,Y) location so that the sparse image can be reconstructed at the host.
PrimeLocate Data Reduction
Windows provides a convenient method for taking advantage of compression by saving images to a “compressed” folder on the hard disk. Windows will dynamically compress the image data as the images are written. An uncompressed TIFF format image stack consisting of 1000 frames of 512 x 512, 16-bit image data without PrimeLocate enabled is 504MB uncompressed, and 346MB when saved to a compressed folder. However, when using PrimeLocate configured for 16 regions of 31×31 pixels, the same file consumes only 54 MB in the compressed folder as the blank-zero valued area will now efficiently compress using the Windows compression algorithm.
When using the maximum 512 regions, more data from the original image is transferred, and this same file is reduced to 257 MB. These results are typical, the exact amount of compression depends on the image data and the number and size of stored regions.
Summary
Computational imaging methods are powerful pieces of software that can greatly assist specific needs, and the Prime camera family comes packed with several pieces of software to ease and improve quantitative scientific imaging. PrimeLocate dynamically evaluates acquired images and reduces the surplus of data generated during high-speed super-resolution imaging, well suited for localization microscopy.
