Introduction
While most fluorescent microscopy involves looking at hundreds or thousands of fluorophores spread throughout a sample, it is sometimes necessary to observe specific molecules or small groups of molecules.
In addition, rather than just detecting the position of fluorophores, some experiments involve analyzing the molecules further, describing the biophysics or kinetic movement of molecules. For all these kinds of experiments there exists a subset of fluorescence microscopy techniques termed single molecule fluorescence microscopy (SMFM). SMFM allows for the investigation of the behavior of single molecules (or small groups) under very strict conditions, which ensures that each molecule is in the same state as any other molecule.
Labeling
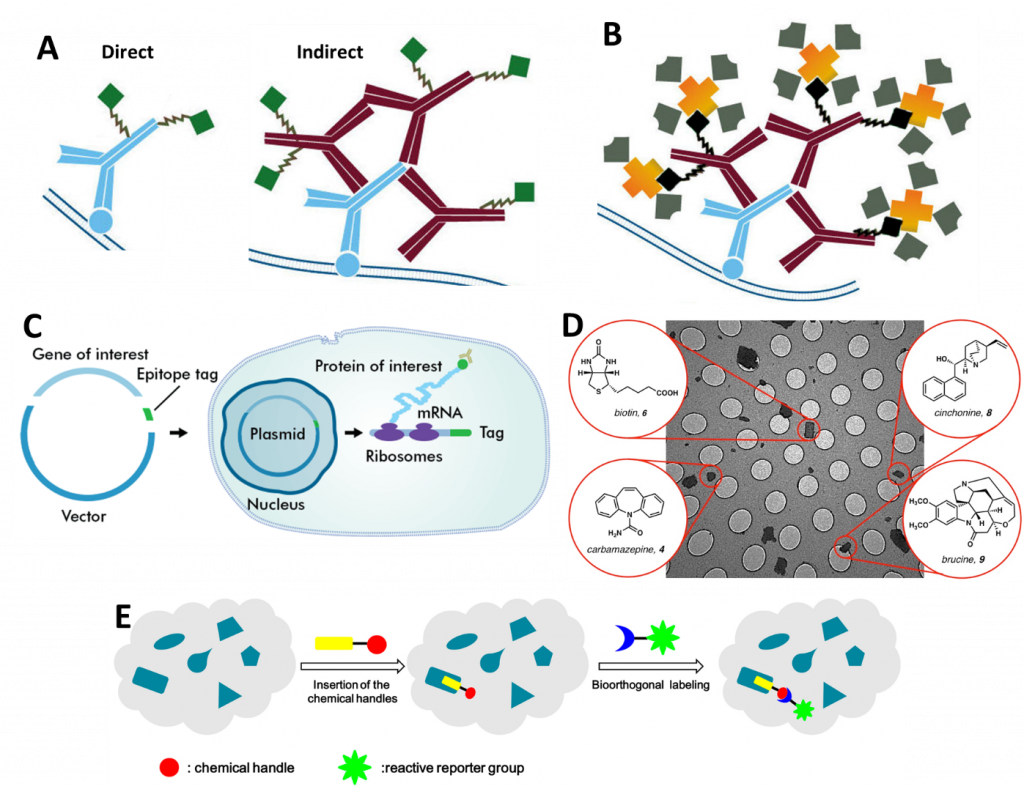
In essence, SMFM involves attaching a fluorophore to a single molecule (unless the molecule is inherently fluorescent already) and then imaging and/or tracking it over time. There are numerous techniques used to attach fluorophores, the main ones being: antibodies (Fig.1A), Biotinylation (Fig.1B), epitopes (Fig.1C), small molecule probes (Fig.1D) and bioorthogonal labeling (Fig.1E), all outlined in Fig.1. For further information, refer to the App Note: Labelling Proteins for Single Molecule Imaging.
As for what type of fluorophore to use, for SMFM fluorophores are required to be:
- Bright, with the ability to strongly absorb excitation light and efficiently fluoresce
- Photostable, so that it does not bleach quickly (except for FRAP) and can be imaged for long periods of time
- Small, so that it does not disrupt the biological activity of the molecule
- Emitting light in the visible region of the spectrum (not infrared or ultraviolet emission), preferably in the region where the camera is most efficient
The most popular fluorophores for SMFM include organic dyes such as fluorescein isothiocyanate (FITC, green fluorescence) and tetramethylrhodamine (TRITC, red fluorescence)), fluorescent proteins (such as green/yellow fluorescent protein (GFP/YFP) and quantum dots (semiconductor particles a few nanometers in size).
It is important to get the optimal fluorophore and labeling method for SMFM as this application is limited by noise, which means the signal-to-noise ratio (SNR) can be low unless the fluorophore can produce a strong enough signal. Even so, a very sensitive camera is necessary to detect the relatively low signal in the high background noise environment, which is worsened if the experiments need to be captured quickly with high framerates (as this lowers the exposure time). A high-speed and highly sensitive camera is an absolute must when performing SMFM.
Techniques
In SMFM there are typically two main categories of experiment: either the single molecule is simply observed and tracked in a system passively, or a force is applied actively to the system, such as physical tension with optical tweezers (for more information, refer to the App Note on Optical Tweezers). The remainder of this article describes passive SMFM techniques.
It should be noted that most conventional microscopy techniques can be applied to image single molecules rather than large groups of molecules, including standard widefield fluorescence, confocal fluorescence microscopy, and super-resolution microscopy (specifically PALM and STORM). Beyond these, there are specific techniques suited for SMFM, such as total internal reflection fluorescence (TIRF), Förster resonance energy transfer (FRET) and fluorescence recovery after photobleaching (FRAP). See our individual app notes on TIRF, FRET, and FRAP for more information.
Examples
Two examples of SMFM are shown in Fig.2 and Fig.3, showing fast single molecule tracking and DNA mapping respectively. Each example showcases single molecule microscopy, with the samples being small populations of a greater whole, and focusing in on specific parts of each sample, be it single molecules or chains of DNA.


Summary
SMFM is used to investigate the biological structure and function of individual molecules. Compared to conventional studies, single molecule imaging reveals far more information about the behavior of molecules that would otherwise be hidden by a large group of molecules. This has resulted in a wealth of new information over a large range of biological processes.
Since the uptake of SMFM in the 1990s, many new techniques have since been developed and embraced by single molecule researchers. With the development of newer, brighter fluorophores and more sensitive scientific cameras, the historical issues of low signal-to-noise in single molecule experiments is becoming less of a limiting factor. This has led to single molecule imaging becoming more and more popular as a fluorescence microscopy technique.
References
Li L., Zhang Z. (2016) Development and Applications of the Copper-Catalyzed Azide-Alkyne Cycloaddition (CuAAC) as a Bioorthogonal Reaction, Molecules, Oct 24, 21 (10).
Qu X., Wu D., Mets L., Scherer N.F. (2004) Nanometer-localized multiple single-molecule fluorescence microscopy, PNAS 101 (31), 11298-11303.
